当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Expanding Extender Substrate Selection for Unnatural Polyketide Biosynthesis by Acyltransferase Domain Exchange within a Modular Polyketide Synthase
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2023-04-14 , DOI: 10.1021/jacs.2c11027
Elias Englund 1, 2 , Matthias Schmidt 1, 3, 4 , Alberto A Nava 3, 5 , Anna Lechner 5, 6 , Kai Deng 1, 7 , Renee Jocic 1, 3 , Yingxin Lin 8 , Jacob Roberts 3, 6 , Veronica T Benites 1, 3 , Ramu Kakumanu 1, 3 , Jennifer W Gin 1, 3 , Yan Chen 1, 3 , Yuzhong Liu 5, 6 , Christopher J Petzold 1, 3 , Edward E K Baidoo 1, 3 , Trent R Northen 1, 9 , Paul D Adams 1, 6, 10 , Leonard Katz 1, 11 , Satoshi Yuzawa 1, 3, 12, 13 , Jay D Keasling 1, 3, 5, 6, 7, 11, 14, 15
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2023-04-14 , DOI: 10.1021/jacs.2c11027
Elias Englund 1, 2 , Matthias Schmidt 1, 3, 4 , Alberto A Nava 3, 5 , Anna Lechner 5, 6 , Kai Deng 1, 7 , Renee Jocic 1, 3 , Yingxin Lin 8 , Jacob Roberts 3, 6 , Veronica T Benites 1, 3 , Ramu Kakumanu 1, 3 , Jennifer W Gin 1, 3 , Yan Chen 1, 3 , Yuzhong Liu 5, 6 , Christopher J Petzold 1, 3 , Edward E K Baidoo 1, 3 , Trent R Northen 1, 9 , Paul D Adams 1, 6, 10 , Leonard Katz 1, 11 , Satoshi Yuzawa 1, 3, 12, 13 , Jay D Keasling 1, 3, 5, 6, 7, 11, 14, 15
Affiliation
|
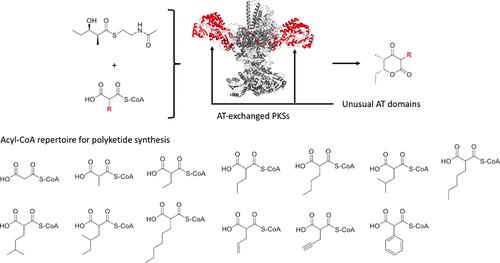
Modular polyketide synthases (PKSs) are polymerases that employ α-carboxyacyl-CoAs as extender substrates. This enzyme family contains several catalytic modules, where each module is responsible for a single round of polyketide chain extension. Although PKS modules typically use malonyl-CoA or methylmalonyl-CoA for chain elongation, many other malonyl-CoA analogues are used to diversify polyketide structures in nature. Previously, we developed a method to alter an extension substrate of a given module by exchanging an acyltransferase (AT) domain while maintaining protein folding. Here, we report in vitro polyketide biosynthesis by 13 PKSs (the wild-type PKS and 12 AT-exchanged PKSs with unusual ATs) and 14 extender substrates. Our ∼200 in vitro reactions resulted in 13 structurally different polyketides, including several polyketides that have not been reported. In some cases, AT-exchanged PKSs produced target polyketides by >100-fold compared to the wild-type PKS. These data also indicate that most unusual AT domains do not incorporate malonyl-CoA and methylmalonyl-CoA but incorporate various rare extender substrates that are equal to in size or slightly larger than natural substrates. We developed a computational workflow to predict the approximate AT substrate range based on active site volumes to support the selection of ATs. These results greatly enhance our understanding of rare AT domains and demonstrate the benefit of using the proposed PKS engineering strategy to produce novel chemicals in vitro.
中文翻译:

通过模块化聚酮合酶内的酰基转移酶域交换扩展非天然聚酮生物合成的扩展底物选择
模块化聚酮合酶 (PKS) 是使用 α-羧基辅酶 A 作为增量底物的聚合酶。该酶家族包含多个催化模块,其中每个模块负责单轮聚酮化合物链延伸。尽管 PKS 模块通常使用丙二酰辅酶 A 或甲基丙二酰辅酶 A 进行链延长,但许多其他丙二酰辅酶 A 类似物用于使自然界中的聚酮化合物结构多样化。以前,我们开发了一种方法,通过在保持蛋白质折叠的同时交换酰基转移酶 (AT) 结构域来改变给定模块的延伸底物。在这里,我们报告了13 种 PKS(野生型 PKS 和 12 种具有异常 AT 的 AT 交换 PKS)和 14 种扩展底物的体外聚酮化合物生物合成。我们的 ~200体外反应产生了 13 种结构不同的聚酮化合物,其中包括几种尚未报道的聚酮化合物。在某些情况下,AT 交换的 PKS 产生的目标聚酮化合物是野生型 PKS 的 100 倍以上。这些数据还表明,大多数不寻常的 AT 结构域不包含丙二酰辅酶 A 和甲基丙二酰辅酶 A,但包含大小等于或略大于天然底物的各种稀有扩展底物。我们开发了一个计算工作流来预测基于活性位点体积的近似 AT 底物范围,以支持 AT 的选择。这些结果极大地增强了我们对稀有 AT 域的理解,并证明了使用所提出的 PKS 工程策略在体外生产新型化学品的好处。
更新日期:2023-04-14
中文翻译:

通过模块化聚酮合酶内的酰基转移酶域交换扩展非天然聚酮生物合成的扩展底物选择
模块化聚酮合酶 (PKS) 是使用 α-羧基辅酶 A 作为增量底物的聚合酶。该酶家族包含多个催化模块,其中每个模块负责单轮聚酮化合物链延伸。尽管 PKS 模块通常使用丙二酰辅酶 A 或甲基丙二酰辅酶 A 进行链延长,但许多其他丙二酰辅酶 A 类似物用于使自然界中的聚酮化合物结构多样化。以前,我们开发了一种方法,通过在保持蛋白质折叠的同时交换酰基转移酶 (AT) 结构域来改变给定模块的延伸底物。在这里,我们报告了13 种 PKS(野生型 PKS 和 12 种具有异常 AT 的 AT 交换 PKS)和 14 种扩展底物的体外聚酮化合物生物合成。我们的 ~200体外反应产生了 13 种结构不同的聚酮化合物,其中包括几种尚未报道的聚酮化合物。在某些情况下,AT 交换的 PKS 产生的目标聚酮化合物是野生型 PKS 的 100 倍以上。这些数据还表明,大多数不寻常的 AT 结构域不包含丙二酰辅酶 A 和甲基丙二酰辅酶 A,但包含大小等于或略大于天然底物的各种稀有扩展底物。我们开发了一个计算工作流来预测基于活性位点体积的近似 AT 底物范围,以支持 AT 的选择。这些结果极大地增强了我们对稀有 AT 域的理解,并证明了使用所提出的 PKS 工程策略在体外生产新型化学品的好处。

































 京公网安备 11010802027423号
京公网安备 11010802027423号